Training and testing data for predicting protein-binding nucleotides were obtained protein-RNA complexes. Listed below are the PDB codes of 542 protein-RNA complexes obtained from the Protein Data Bank (PDB) solved by X-ray crystallography with a resolution of 3.0 Å or better. The 542 protein-RNA complexes contain 439 RNA sequences and 376 protein sequences.
| 1A34 | |1A9N | |1AQ3 | |1AQ4 | |1ASY | |1ASZ | |1AV6 | |1B23 | |1B2M | |1B7F | |1BMV | |1C0A | |1C9S | |1CVJ | |1CX0 | |1DDL |
| 1DI2 | |1DRZ | |1DUL | |1E7K | |1EC6 | |1EFW | |1EUY | |1EXD | |1F8V | |1FFY | |1FXL | |1G2E | |1G59 | |1GAX | |1GSG | |1GTF |
| 1GTN | |1GTR | |1GTS | |1H2C | |1H2D | |1H3E | |1H4Q | |1H4S | |1HQ1 | |1I5L | |1IL2 | |1IVS | |1J1U | |1JBR | |1JBS | |1JBT |
| 1JID | |1K8W | |1KNZ | |1KQ2 | |1L9A | |1LNG | |1M5K | |1M5O | |1M5P | |1M5V | |1M8V | |1M8W | |1M8X | |1M8Y | |1N1H | |1N35 |
| 1N38 | |1N77 | |1N78 | |1NB7 | |1O0B | |1O0C | |1OOA | |1PGL | |1PVO | |1QF6 | |1QRS | |1QRT | |1QRU | |1QTQ | |1QU2 | |1QU3 |
| 1R3E | |1R9F | |1RC7 | |1RLG | |1RPU | |1S03 | |1SDS | |1SER | |1SI3 | |1SJ3 | |1SJ4 | |1SJF | |1TFW | |1TTT | |1U0B | |1U1Y |
| 1URN | |1UTD | |1UTF | |1UTV | |1UVI | |1UVJ | |1UVK | |1UVL | |1UVM | |1UVN | |1VBX | |1VBY | |1VBZ | |1VC0 | |1VC6 | |1VC7 |
| 1VFG | |1WMQ | |1WNE | |1WPU | |1WRQ | |1WSU | |1XOK | |1Y39 | |1YTU | |1YTY | |1YVP | |1YYK | |1YYO | |1YYW | |1YZ9 | |1ZBH |
| 1ZDH | |1ZDI | |1ZDJ | |1ZDK | |1ZE2 | |1ZH5 | |1ZHO | |1ZJW | |1ZL3 | |1ZSE | |2A1R | |2A8V | |2AB4 | |2ANN | |2ANR | |2ASB |
| 2ATW | |2AZ0 | |2AZ2 | |2AZX | |2B2D | |2B3J | |2BBV | |2BGG | |2BH2 | |2BNY | |2BQ5 | |2BS0 | |2BS1 | |2BTE | |2BU1 | |2BX2 |
| 2C4Q | |2C4Y | |2C4Z | |2C50 | |2C51 | |2CSX | |2CT8 | |2CV0 | |2CV1 | |2CV2 | |2DB3 | |2DLC | |2DR2 | |2DR5 | |2DR7 | |2DR8 |
| 2DR9 | |2DRA | |2DRB | |2DU3 | |2DU4 | |2DVI | |2DXI | |2E9R | |2E9T | |2E9Z | |2EC0 | |2EZ6 | |2F8K | |2F8S | |2FK6 | |2FMT |
| 2FZ2 | |2G4B | |2GIC | |2GJW | |2GXB | |2HVY | |2HW8 | |2HYI | |2I91 | |2IX1 | |2IZ9 | |2IZM | |2IZN | |2J0S | |2JEA | |2JLU |
| 2JLV | |2JLW | |2JLX | |2JLY | |2JLZ | |2NUE | |2NUF | |2NUG | |2NZ4 | |2OIH | |2OJ3 | |2OZB | |2PJP | |2PLY | |2PO1 | |2PXB |
| 2PXD | |2PXE | |2PXF | |2PXK | |2PXL | |2PXP | |2PXQ | |2PXT | |2PXU | |2PXV | |2PY9 | |2Q66 | |2QUX | |2R7R | |2R7T | |2R7V |
| 2R7W | |2R7X | |2R8S | |2RD2 | |2RE8 | |2RFK | |2UWM | |2V3C | |2VNU | |2VOD | |2VON | |2VOO | |2VOP | |2VPL | |2X1A | |2X1F |
| 2XD0 | |2XDB | |2XGJ | |2XLI | |2XLJ | |2XLK | |2XNR | |2XS2 | |2XS5 | |2XS7 | |2XZO | |2Y8W | |2Y8Y | |2Y9H | |2YJY | |2YKG |
| 2ZH1 | |2ZH2 | |2ZH3 | |2ZH4 | |2ZH5 | |2ZH6 | |2ZH7 | |2ZH8 | |2ZH9 | |2ZHA | |2ZI0 | |2ZKO | |2ZM5 | |2ZUE | |2ZUF | |2ZXU |
| 2ZZM | |2ZZN | |3ADB | |3ADC | |3ADD | |3ADL | |3AEV | |3AF6 | |3AGV | |3AHU | |3AKZ | |3AM1 | |3AMT | |3AVT | |3AVU | |3AVW |
| 3AVX | |3AVY | |3B0U | |3BOY | |3BSB | |3BSN | |3BSO | |3BSX | |3BT7 | |3BX2 | |3BX3 | |3CUL | |3CUN | |3D2S | |3DD2 | |3DH3 |
| 3EGZ | |3EPH | |3EQT | |3ER9 | |3EX7 | |3FHT | |3FOZ | |3FTE | |3FTF | |3G0H | |3G8T | |3G9C | |3G9Y | |3GIB | |3GPQ | |3H5X |
| 3H5Y | |3HAX | |3HHN | |3HJW | |3HL2 | |3HSB | |3I5X | |3I5Y | |3I61 | |3I62 | |3IAB | |3ICE | |3IE1 | |3IEM | |3IEV | |3IRW |
| 3K49 | |3K5Q | |3K5Y | |3K5Z | |3K61 | |3K62 | |3K64 | |3KLV | |3KMQ | |3KMS | |3KNA | |3KOA | |3KS8 | |3L25 | |3L26 | |3L3C |
| 3LQX | |3LRN | |3LRR | |3LWO | |3LWP | |3LWQ | |3LWR | |3LWV | |3M7N | |3M85 | |3MDG | |3MDI | |3MJ0 | |3MOJ | |3MQK | |3MUM |
| 3MUR | |3MUT | |3MXH | |3NCU | |3NDB | |3NL0 | |3NMA | |3NMR | |3NMU | |3NNA | |3NNC | |3NNH | |3NVI | |3O3I | |3O6E | |3O7V |
| 3O8C | |3O8R | |3OG8 | |3OL6 | |3OL7 | |3OL8 | |3OL9 | |3OLB | |3OUY | |3OV7 | |3OVA | |3OVB | |3OVS | |3P6Y | |3PEW | |3PEY |
| 3PF4 | |3PF5 | |3PTX | |3PU4 | |3Q0L | |3Q0M | |3Q0N | |3Q0O | |3Q0P | |3Q0Q | |3Q0R | |3Q0S | |3QG9 | |3QGB | |3QGC | |3QJJ |
| 3QJL | |3QRP | |3QSU | |3R2C | |3R2D | |3R9W | |3R9X | |3RC8 | |3RER | |3RTJ | |3RW6 | |3SIU | |3SN2 | |3SNP | |3SQW | |3SQX |
| 3T3O | |3T5N | |3T5Q | |3TMI | |3TRZ | |3TS0 | |3TS2 | |3U2E | |3U4M | |3U56 | |3UCU | |3UCZ | |3UD4 | |3UMY | |3V6Y | |3V74 |
| 3VJR | |3VNV | |3VYX | |3VYY | |3ZC0 | |3ZD6 | |3ZD7 | |3ZGZ | |4AFY | |4AL5 | |4AL6 | |4AL7 | |4ALP | |4AM3 | |4AQ7 | |4ARC |
| 4ARI | |4AS1 | |4ATO | |4AY2 | |4B3G | |4BA2 | |4BPB | |4E78 | |4ED5 | |4EI1 | |4EI3 | |4EJT | |4ENN | |4ERD | |4F02 | |4F3T |
| 4FTB | |4FVU | |4G0A | |4G9Z | |4GCW | |4GHA | |4GHL | |4GV3 | |4GV6 | |4GV9 | |4H5P | |4HOR | |4HOS | |4HOT | |4HT8 | |4HT9 |
| 4I67 | |4IFD | |4IG8 | |4ILL | |4IQX | |4J1G | |4J7L | |4J7M | |4JNG | |4JVY | |4JXX | |4JXZ | |4JYZ | |4JZU | |4JZV | |4K4S |
| 4K4T | |4K4U | |4K4W | |4K4X | |4K4Z | |4K50 | |4KRE | |4KRF | |4KXT | |4L8R | |4LG2 | |5MSF | |6MSF | |7MSF | | |
Two datasets were constructed for training two prediction models: (1) training set for the prediction model that uses both RNA and protein sequences, and (2) training set for the prediction model that uses RNA sequence alone.
| Dataset | total number of protein-binding nucleotides in RNA sequences | total number of non-binding nucleotides in RNA sequences | Download |
| Training set for the prediction model that uses both RNA and protein sequences | 2,716 | 6,432 | Link |
| Training set for the prediction model that uses RNA sequence alone | 2,189 | 4,588 | Link |
Two independent datasets were constructed with sequences that were not used in training. (1) test set for the prediction model that uses both RNA and protein sequences, and (2) test set for the prediction model that uses RNA sequence alone.
| Dataset | total number of protein-binding nucleotides in RNA sequences | total number of non-binding nucleotides in RNA sequences | Download |
| Test set for the prediction model that uses both RNA and protein sequences | 1,848 | 4,631 | Link |
| Test set for the prediction model that uses RNA sequence alone | 1,795 | 4,235 | Link |
A binding site should be involved in at least one of the following interactions between RNA and protein: hydrogen bonds, water bridges and hydrophobic interactions. [Click here] for detailed information.
We define a protein–RNA binding site by three types of interactions: hydrogen bonds, water bridges and hydrophobic interactions. A nucleotide involved in at least one of the interactions is classified as a protein-binding site in RNA. For each of the 542 protein–RNA complexes, we obtained the three types of interactions from NPIDB and incorporated them into the RNA sequences of protein–RNA complexes.
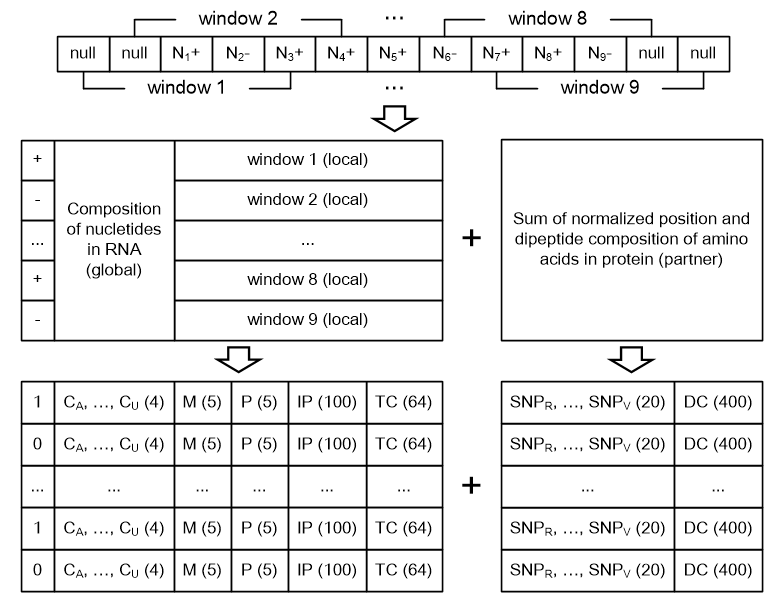
| Feature | Type | Size | Meaning |
| C | Global | 4 | Nucleotide compositions |
| M | Local | 1 * window size | Mass of nucleotides |
| P | Local | 1 * window size | pKa value of nucleotides |
| IP | Local | 20 * window size | Interaction propensity of nucleotide triplets |
| TC | Local | 64 | Triplet compositions of the RNA sequence |
| SNP | Partner | 20 | Sum of the normalized position of amino acids |
| DC | Partner | 400 | Dipeptide compositions of the protein sequence |
normalized position: position number of an amino acid divided by the sequence length
Example of encoding a RNA sequence of 9 nucleotides by a sliding window of size 5
| 10-fold cross validation |
| Model | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| RP | 87.4% | 84.4% | 85.3% | 70.3% | 94.1% | 0.68 |
| RaP | 87.1% | 73.5% | 77.9% | 61.0% | 92.3% | 0.57 |
| Independent test |
| Model | Sensitivity | Specificity | Accuracy | PPV | NPV | MCC |
| RP | 72.8% | 71.7% | 72.0% | 50.7% | 86.9% | 0.41 |
| RaP | 68.4% | 70.6% | 69.9% | 49.6% | 84.0% | 0.36 |
RP: prediction of protein-binding sites in RNA from protein and RNA sequences
RaP: Prediction of protein-binding sites in RNA from RNA sequence alone
PPV: positive predictive value; NPV: negative predictive value; MCC: Matthews correlation coefficient